Research Projects
|
Genetics of Dietary Adaptation and Host-Microbiota Interactions
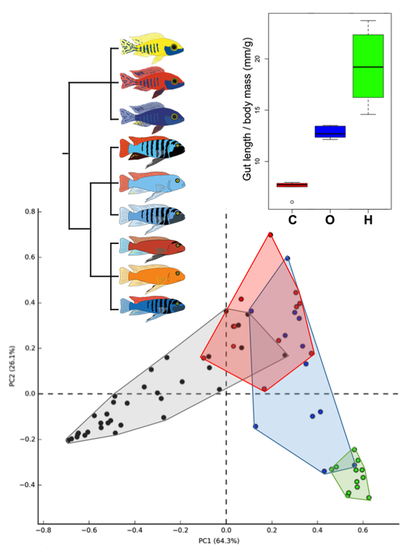
Maladaptive diets are the leading contributor to morbidity and mortality worldwide. Disease outcomes stemming from both undernourishment and overconsumption represent evolutionary discordance: the human genome has been unable to evolve to match rapid dietary changes brought on by overpopulation and the agricultural and industrial revolutions. If we reveal pathways of genetic evolution taken by other vertebrates as they adapted to diverse diets, we should identify pathways for therapeutic intervention for pathological conflicts in humans brought on by maladaptive diets. East African cichlid fish species have recently evolved diverse dietary adaptations, providing powerful comparative experimental strategies to understand the genetic basis of dietary response. We have used genomic, metagenomic, and developmental biology techniques to demonstrate divergence in gastrointestinal morphology and function among species representing different trophic levels. Ongoing research includes interspecific hybrid crosses and comparative genomic strategies to reveal the genes involved in species-specific gut morphology and microbiota. Funding: The Arnold and Mabel Beckman Foundation Beckman Young Investigator Award, “A Novel Genetic Model of Dietary Response and Host-Microbiota Interactions” |
|
Polygenic Sex Determination
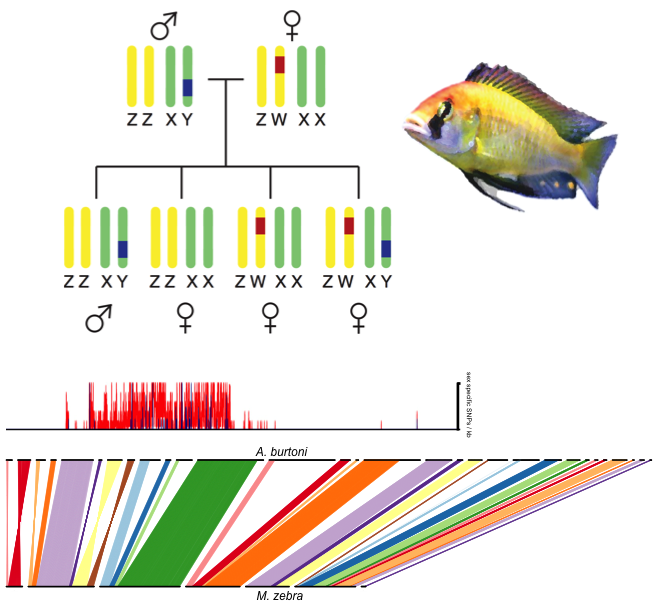
Historically, genetic sex determination research has largely focused on a handful of single-factor, chromosomal sex determination systems. More recently, genetic sex determination has been mapped in a broader set of species, revealing a diverse catalog of sex determination genes in a variety of chromosomal contexts. Polygenic sex determination (PSD) has also been confirmed in multiple taxa, where multiple genetic factors direct sexual development. PSD challenges traditional thinking regarding genetic sex determination systems, while providing powerful strategies to explore the evolution and development of sex determination, including comparisons of siblings with multiple genetic modes of sex determination. In other words, while individuals of species with PSD develop as male or female, several genetic types of males and females may exist. Multiple cichlid fish species use PSD to determine male vs. female development, providing models to understand the evolution and development of genetic sex determination. Our current work aims to provide the most comprehensive analysis of a PSD system to date, using the species Astatotilapia burtoni as a model system. We are working to reveal novel genes and gene networks driving gonadal differentiation and development, while producing important genome resources for A. burtoni, a model for neural and behavioral biology. Funding: The National Science Foundation, “Complex Evolutionary Genetics of Gonadal Differentiation in a Classic Model of Behavior, Astatotilapia burtoni” |
|
Genetics of Species-Specific Behavior
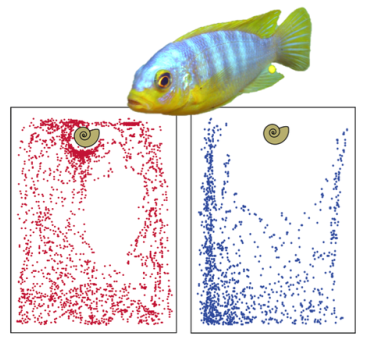
The evolution of vertebrate behavior has long fascinated the scientific community, but an understanding of the genetic basis of species-specific behavior has been limited by challenges associated with quantification of complex behaviors, as well as a lack of genetically tractable model systems. We are developing Lake Malawi cichlids as a system to identify the genetic basis of behavioral divergence, by comparing closely related sister species with distinct behaviors. Combining detailed modern phenotypic analysis in representative species in the lab, genetic mapping in hybrid crosses between these species, and comparative genomic analysis across the adaptive radiation of East African cichlids, our ultimate goal is to reveal the genetic architecture underlying divergence in complex behaviors, while providing rich evolutionary context for our findings. Funding: NC State Genetics Program Provost Research Award (to Emily Moore) "The Genetic Basis of Species-Specific Exploratory behaviors in African Cichlid Fishes"; NC State College of Sciences |
|
Collaborative Research
Genetics of Trophic Morphology and Pigmentation - Craig Albertson, UMass-Amherst; Brian Langerhans, NC State University Sex Determination in Cichlids - Russell Fernald, Stanford University; Bethany Dumont, NC State University Ear and Armpit Microbiota and Human Genetic Variation - Julie Horvath, NC Museum of Natural Sciences & NC Central University; Rob Dunn, NC State University Dietary Adaptation in Astyanax - Masato Yoshizawa, University of Hawai’i - Manoa Poison Frog Transcriptomes - Kyle Summers, East Carolina University Vole Vaginal Microbiota - Lisa McGraw, NC State University Sex Determination in Marine Fishes - Martha Burford-Reiskind, Harry Daniels, Ben Reading, NC State University; Julia Stevens, NC Museum of Natural Sciences Kudzu Bug Microbiota - Marcé Lorenzen, NC State University |